This post was originally published on Discoveries, the blog of Boston Children’s Hospital.
Glioblastoma, a cancer that arises in the brain’s supporting glial cells, is one of the worst diagnoses a child can receive. The grade IV, highly malignant tumor aggressively infiltrates healthy brain tissue, and most children die of the disease within one to two years of diagnosis, similar to adults.
“The current approach is surgical removal, radiation, and then some kind of chemotherapy, but there is no chemotherapy that improves survival,” says Mariella Filbin, MD, PhD, a pediatric neuro-oncologist at Dana-Farber/Boston Children’s Cancer and Blood Disorders Center. “It’s a very vicious tumor that we haven’t gotten a handle on.”
Filbin co-led a study, published online last week in Cell, that made some surprising discoveries about this poorly understood tumor. Regardless of what genetic mutation causes glioblastoma, it can readily shift among four distinct cell types, each of which may need to be targeted separately.
Profiling a tumor, cell by cell
Usually diagnosed in school-age children, childhood glioblastoma outwardly looks like the adult glioblastoma that killed John McCain, Ted Kennedy, and Beau Biden. But genetically, it is completely different: like many childhood cancers, its various genetic mutations tend to hijack normal pathways of early brain tissue development, which regulate cell functions.

In both children and adults, glioblastoma can take many forms – hence the cancer’s full name, glioblastoma multiforme. But it had been thought that within a given patient, all tumor cells are the same.
That turns out not to be true. The tumor is a mosaic.
In a group effort — “no single lab can do this alone” — Filbin teamed up with Cyril Neftel in the lab of Mario Suvà, MD, PhD, at Massachusetts General Hospital and the Broad Institute; Julie Laffy in the lab of Itay Tirosh, PhD of the Weizmann Institute of Science; and Toshiro Hara, PhD of the Salk Institute. These researchers, all co-first authors on the paper, analyzed tumors from eight children and 20 adults, going cell by cell.
In all, they profiled more than 24,000 cells, sequencing the RNA of each cell. While DNA is hard-wired, the RNA “transcriptome” is more like software, providing a readout of what genes are turned on and what proteins the cell is making — revealing what the cell is doing and what state it’s in.
Applying novel technologies to these vicious tumors give us an opportunity to understand what drives them at a whole new level that was unthinkable a few years ago.
“If you look at single cells, they’re very different from one another,” says Filbin. “Some are more immature, stem-like cells, some are differentiating cells, and some are mature cells that are not even dividing anymore.”
I’ll morph to someone else
The team distinguished four distinct tumor cell types. Each type closely resembled different types of normal developing brain cells: neural progenitor cells, oligodendrocyte progenitor cells, astrocytes, and mesenchymal cells. The proportions of the different cell types varied according to the genetic defect driving the patient’s cancer (increased copies of PDGFRA, CDK4 or EGFR, or alterations of NF1). But every patient had all four tumor cell types.
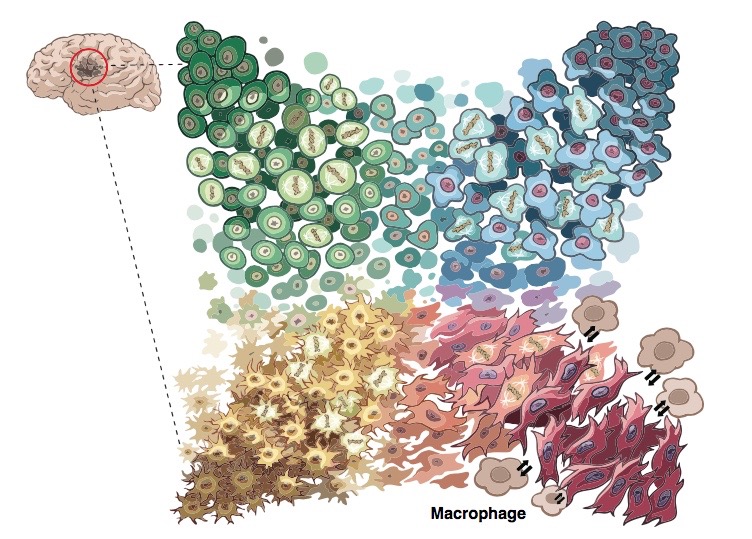
More ominously, the four cell types could all change into each other. The study found that glioblastomas are highly “plastic” — their cell types readily morphing over time and when exposed to cancer treatments. When the researchers injected any of the four types of patient tumor cells into mice, they all formed tumors containing all four cell types.
“This explains the failures of single-target drugs,” says Filbin. “The tumors can ‘become’ something else and escape our drug therapies — and it’s very easy for them.”
Uncovering cancer-cell ‘hierarchies’
Recently, Filbin and her colleagues used single-cell RNA sequencing to profile another deadly tumor called diffuse midline glioma, also known as diffuse intrinsic pontine glioma (DIPG). In tumors caused by one specific mutation, they found a rogue immature type of cell that constantly divided and proliferated much like stem cells — fueling the tumor’s spread.

Unlike glioblastoma, DIPG cells seemed to morph in just one direction: a few rogue cells were able to differentiate into more mature cells. That offers a new approach to explore: getting these tumor-propagating cells to mature and become less of a threat. Filbin thinks other tumors might follow this “unidirectional hierarchy.”
“The younger the child, the more immature the tumors, and the more unidirectional we think they will be, because they resemble early development so much,” she says. “Older children may be more likely to follow a glioblastoma pattern.”
Treating glioblastoma: The plot thickens
As for glioblastoma, the new findings indicate a need for combination therapies that target all four tumor states at once. Filbin and her colleagues plan to try different combinations of gene editing approaches and drugs aimed at changing cell state. Such drugs could block molecular pathways that keep tumor cells in an immature state, or target the packaging of DNA into chromatin that determines what RNA is transcribed.
“Applying novel technologies to these vicious tumors give us an opportunity to understand what drives them at a whole new level that was unthinkable a few years ago,” says Filbin. “The time is ripe to take these findings and finally translate them into novel therapies.”
Tirosh and Suvà were co-senior investigators on the glioblastoma study, whose supporters included numerous foundations, the American Cancer Society, the National Cancer Institute, and the National Institutes of Health. Neftel, Filbin, Hara and many coauthors are also affiliated with the Broad Institute of Harvard and MIT. The authors declare no competing interests. See the paper for a complete list of funders and authors.
